Global sequencing and surveillance capacity for SARS-CoV-2 must be strengthened and combined with multidisciplinary studies of infectivity, virulence and immune escape, in order to track the unpredictable evolution of the ongoing COVID-19 pandemic.
In June 2020, the World Health Organization (WHO) SARS-CoV-2 evolution working group was established to track SARS-CoV-2 variants and their specific genetic changes and to monitor viral characteristics and their impact on medical and non-medical countermeasures, including vaccines against COVID-19. In November 2021, this working group transitioned to a formal WHO Technical Advisory Group on Virus Evolution (TAG-VE), with the aim of developing and implementing a global risk-monitoring framework for SARS-CoV-2 variants, based on a multidisciplinary approach that includes in silico, virological, clinical and epidemiological data.
Tracking variants
The main role of the TAG-VE is to function as an integrative forum for the exchange of information from global surveillance and research studies to monitor early warning signals, and to perform timely assessment of the need for public health action in response to emerging variants1. It uses a Delphi consensus method to establish which emerging variants are considered variants of interest (VOIs) or variants of concern (VOCs) (definitions, ref. 2). To avoid stigmatizing countries that first identify and report variants, a naming scheme that follows WHO guidelines3 and assigns Greek letters to VOIs and VOCs was adopted for global discourse in June 2021 (ref. 4). As of 26 March 2022, eight VOIs and five VOCs had been designated, and they have been further described as previously or currently circulating variants to reflect changes in their epidemiology over time2,5.
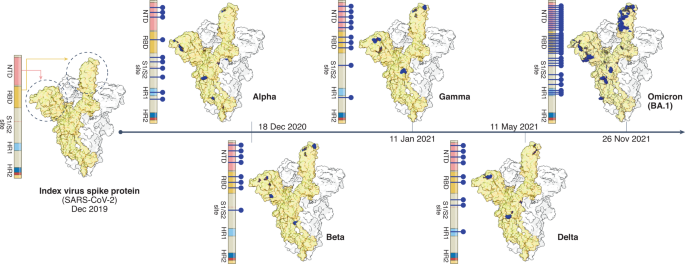
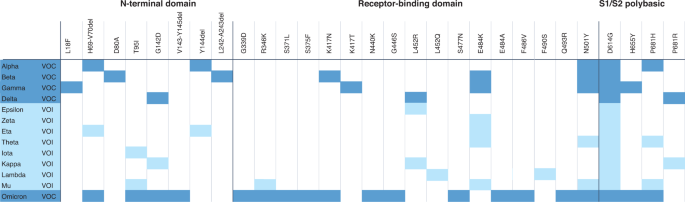
The first available data on emerging variants that are assessed by TAG-VE are viral sequences and their associated metadata shared on publicly accessible genetic sequence databases (e.g., GISAID, Genbank, the European Nucleotide Archive, and the DNA Database of Japan). Because all current VOIs and VOCs originated from ancestral variants, available sequences are still compared with that of the 2019 index virus (GISAID accession ID: EPI_ISL_402124), with the primary focus on regions of the genome that are known to encode important viral or immunity- or infection-associated proteins. Mutations in the gene encoding the spike protein are given highest priority, as they have the highest probability of being clinically important. The spike protein contains the receptor-binding domain that is essential for docking of the virus on host cells, as well as major determinants of both viral transmissibility (for example, the polybasic cleavage site) and antigenic makeup (the N-terminal domain and receptor-binding domain). However, mutations in genes encoding proteins other than spike have also been found to be important and should not be neglected6. The constellation of mutations detected in any emerging variant is compared with a list of annotated mutations that are suspected or known to have a role in one or more of the viral characteristics included in the definitions of VOI and VOC: transmissibility, immune escape, disease severity, detectability and susceptibility to available treatments. Figure 1 depicts the evolution of SARS-CoV-2 and the variant-defining sets of amino acid substitutions in spike seen in VOCs. Thus far, researchers have studied more than two dozen spike proteins, including those with phenotypes that confer immune escape (Fig. 2). The list of mutations and their associated phenotypic impact, directly demonstrated in laboratory studies or inferred through the use of in silico methods, is updated regularly on the basis of timely research presentations from the WHO partner network, as well as searches of preprints and peer-reviewed published literature2. This annotated list of mutations has proven useful, as the same mutations have independently arisen in different lineages of SARS-CoV-2, strongly suggestive of viral adaptation to the human host and selective pressure from population immunity.
Spike structure of the index virus (far left), together with key domains (highlighted in different colors), including the N-terminal domain (NTD), the receptor-binding domain (RBD), the S1–S2 junction (including the polybasic cleavage site), and the heptapeptide repeat domains (HR1 and HR2). For the five VOCs (middle and right), blue lines with dots indicate location of variation in each domain, and blue dots in adjacent protein structures indicate location of variation in that structure. The VOCs are plotted chronologically according to date of identification.
Notably, the detection of a new constellation of mutations in a variant does not necessarily translate into an increased public health threat, which suggests that some mutations, or combinations of mutations, may impart a fitness cost, rather than benefit. For example, the VOI Theta had a constellation of mutations that alerted scientists because it included top-ranked mutations such as E484K, N501Y, D614G and P681H in spike (Fig. 2), but its spread remained very limited. To establish whether a new variant poses a serious threat, TAG-VE looks for early epidemiological signals of spread and clinical signals derived from surveillance data or specific studies. This includes assessing measures of how quickly cases are increasing and in what geographical areas and population subgroups a variant emerges and spreads, as well as changes to disease severity indicators, such as hospitalization. Careful consideration is also given to the assessment of evidence of relative transmissibility, compared with that of other circulating variants, including secondary attack rates observed in household transmission studies. Finally, the assessment of the threat posed by a variant also needs to consider vaccine- or infection-derived population immunity, which has progressively grown to high levels in many countries, and has also become more complex in terms of different permutations of hybrid immunity, which refers to the immunity elicited by both vaccination and infection7. Therefore, studies that look at the prevalence of re-infections or vaccine breakthroughs are also reviewed, where available.
Access to samples
The experience gained since the launch of the virus evolution working group, which later became the TAG-VE, has shown that the speed with which new SARS-CoV-2 variants spread can outpace the current ability to assess their threat. Variant sequence determination has now become part of the global surveillance, and systematic phenotypic characterization of emerging variants is a critical component that needs to be added to the surveillance core toolbox. It is therefore important to define gaps that need to be addressed to enable better and more timely responses to new variant threats.
Although genomic indicators for transmissibility and immune escape of VOCs are conceptually relatively straightforward to assess, the wide diversity of assays, the lack of centralized biobanks of viruses and clinical specimens, the difficulties in international shipping of materials, and the globally fragmented funding landscape make the comparability of data amassed in real time challenging. To improve the global capacity to rapidly perform phenotypic characterization, viral isolates of emerging variants should be promptly generated and shared to enable researchers in different laboratories to work with viruses that carry the same constellation of mutations. To address this challenge, the WHO is developing a bio-hub system and recently established a BioHub facility8. Its aim is to offer a reliable, safe and transparent mechanism for WHO Member States to voluntarily share novel biological materials, without replacing or competing with existing systems such as EVAg (European Virus Archive Global) and the US National Institutes of Health BEI (Biodefense and Emerging Infections) repository.
Sequence data
Another important challenge is the availability, representativeness and quality of genetic sequence data, which vary depending on sample quality and processing and the experience of the researchers in handling samples, and the platforms, protocols and analytical tools used, and access to all of these. This challenge is exemplified in the large bias in the volume of sequences contributed by a small number of countries to global databases, as well as the release of sequences with low-confidence genomic regions and gaps. For example, amplicon-based methods are sensitive to mutations, which leads to amplification errors that can lead to sequence errors or poor-quality sequence in certain regions of the genome. The consequences of viral diversification call for sustained, if not enhanced, investment from governments in the capacity of reference laboratories to match the global public health demand for high-quality sequences and viral characterization for SARS-CoV-2, which can then be used for future public health threats posed by any infectious disease.
Verification of sequence quality through analysis of raw reads is a critical quality check that becomes even more critical for the detection of recombination between SARS-CoV-2 genomes. Verified detection of SARS-CoV-2 circulating recombinant forms since the Omicron VOC emerged has increased, most likely because of increased availability of genomic surveillance, as well as natural factors. The larger number of lineage-defining mutations in Omicron makes the detection of recombinant forms easier, and re-infection with immune-escape variants increases the chances of co-infection and therefore recombination, especially if two variants co-circulate in the community at high levels. Although it is impossible to predict whether circulating recombinant forms with specific genomic breakpoints may become more transmissible, for a new variant to spread widely, it must inherit traits from the parental viruses that provide a selective advantage.
Infectivity and virulence
Immune escape of VOCs can be shown through live virus-neutralization studies, with results from different laboratories showing similar results. In contrast, there are many methodological gaps in how to assess infectivity and virulence in vitro, ahead of clinical and epidemiological data. Some critical sites and amino acid substitutions have been identified, but this list is not comprehensive; these sites influence determinants of infectivity such as receptor binding, cleavage of the S1 and S2 domains of the spike protein, and cellular entry (Table 1). In vivo animal studies can help elucidate specific features that may be difficult to estimate from epidemiologic data, such as assessing cross-neutralization between variants, which is challenging in humans, as, unlike animal models, they may have unknown prior exposures. Animal models can also help in the assessment of virulence without the confounding effects of background immunity, but it remains to be seen how animal data correlate with disease severity in humans9,10,11,12.
Disease severity in the clinical setting can be especially difficult to determine rapidly and accurately because of challenges posed by ascertainment, inclusion, quality of care, and confounding and colliding biases from empirical observational studies. A key role will be served by the interoperability of electronic health records, which should allow easy linkage of sequencing data with patients’ clinical records. Unbiased and systematic collection of clinical and epidemiological data with comprehensive biological sampling and rigorous characterization of virulence and correlates of protection remains the gold standard for variant threat assessment. The generation of such data from well-designed studies across diverse healthcare settings and different geographies is expected to remain challenging, especially once the response to COVID-19 is de-escalated in the post-acute phase of the pandemic. Integration of such studies into newly established routine surveillance system work plans may help ensure that clinical and epidemiological studies continue in the future.
Until November 2021, the emergence of VOCs such as Alpha and Delta was associated mainly with an increase in transmissibility, most likely driven by viral adaptation to the human host, with modest degrees of immune escape13. The emerging evidence on Omicron variants suggests that immune escape was a substantial driver for its observed displacement over Delta, and its selective advantage was driven by increasing population immunity, in addition to its increased transmissibility14. Although the lower virulence of Omicron was most likely a chance event, the preservation of cellular immunity that protects against severe disease is likely to be a recurring theme15,16. In addition, hybrid immunity in people who have experienced a breakthrough infection seems to broaden immune responses, which suggests that population immunity may be able to tolerate considerable continued evolution of SARS-CoV-217,18. However, because transmission and virulence are uncoupled for SARS-CoV-2, it cannot be assumed that the next variant will be less virulent. Future variants may have virulence similar to or higher or lower than that of Omicron.
An early warning system
A recent retrospective analysis suggested that some of the key variant-defining mutations could potentially have been detected much earlier, which shows the importance of early warning bioinformatics tools from globally shared data, an approach in its infancy19. The identification of key mutations could then trigger a subsequent virus-, variant- or mutation- characterization pipeline. Machine learning algorithms to determine the potential impact of key mutations are being developed and validated, but full assessment of variants still requires epidemiological data and in vivo and/or in vitro experiments, as a VOC cannot be ascertained solely through genomic data.
The future of the pandemic is difficult to predict, for several reasons. First, unlike variants of other respiratory viruses, such as human influenza viruses, SARS-CoV-2 VOCs have so far not emerged from the most recently dominant circulating virus20. Second, chronically infected patients can allow the acceleration of intra-host evolution21. Third, a wide number of susceptible mammals, including cervids, mustelids and rodents, may act as secondary reservoirs with the potential for reverse zoonoses22. Fourth, too much of the world’s vulnerable population remains unvaccinated at present, and current vaccines are suboptimal in preventing transmission. Given these challenges in predicting the evolution of the virus, targeted surveillance of suspected high-risk populations, such as chronically infected patients, and improving detection systems, such as a One Health approach, which also includes wastewater and animal surveillance, are critical for the early detection of future variants.
The evolving virus and the uncertainty of predicting the trajectory of the pandemic call for strengthened surveillance and continued monitoring of SARS-CoV-2. The TAG-VE will continue to critically appraise state-of-the-art methodologies for predicting further evolution of SARS-CoV-2 and will continue to rapidly determine the threat levels posed by new variants. The pandemic is not over, and SARS-CoV-2 is spreading at a high level globally. Now is the time to enhance global sequencing capacities, focusing on widening coverage to include previous geographical and population blind spots, and to build a global consensus toward continued concerted multidisciplinary efforts, under the leadership of the WHO R&D Blueprint for action to prevent epidemics, to track and assess the threat posed by future SARS-CoV-2 variants.
References
-
World Health Organization. https://www.who.int/publications/m/item/terms-of-reference-for-the-technical-advisory-group-on-sars-cov-2-virus-evolution-(tag-ve) (18 October 2021).
-
World Health Organization. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed 15 May 2022).
-
World Health Organization. https://apps.who.int/iris/bitstream/handle/10665/163636/WHO_HSE_FOS_15.1_eng.pdf (May 2015).
-
Konings, F. et al. New Microbiol. 6, 821–823 (2021).
-
World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed 15 May 2022).
-
Syed, A. M. et al. Science 374, 1626–1632 (2021).
-
Bergeri, I. et al. Preprint at MedRxiv https://doi.org/10.1101/2021.12.14.21267791 (2020).
-
World Health Organization. https://www.who.int/initiatives/who-biohub (accessed 15 May 2022).
-
Davies, N. G. et al. Nature 593, 270–274 (2021).
-
Twohig, K. A. et al. Lancet Infect. Dis. 22, 35–42 (2022).
-
Munster, V. J. et al. Sci. Adv. 7, eabj3627 (2021).
-
Muñoz-Fontela, C. et al. PLoS Pathog. 18, e1010161 (2022).
-
Oude Munnink, B. B. et al. Nat. Med. 27, 1518–1524 (2021).
-
Viana, R. et al. Nature 603, 679–686 (2022).
-
Gao, Y. et al. Nat. Med. 28, 472–476 (2022).
-
Keeton, R. et al. Nature 603, 488–492 (2022).
-
Kitchin, D. et al. Cell Rep. Med. 3, 100535 (2022).
-
He, W. et al. Preprint at BioRxiv https://doi.org/10.1101/2021.09.08.459480 (2021).
-
Maher, M. C. et al. Sci. Transl. Med. 14, eabk3445 (2022).
-
Jones, S. et al. Sci. Rep. 9, 14690 (2019).
-
Corey, L. et al. N. Engl. J. Med. 385, 562–566 (2021).
-
Oude Munnink, B. B. et al. Science 371, 172–177 (2021).
-
Grabowski, F., Preibisch, G., Giziński, S., Kochańczyk, M. & Lipniacki, T. Viruses 13, 392 (2021).
-
McCallum, M. et al. Cell 184, 2332–2347.e16 (2021).
-
Wang, P. et al. Nature 593, 130–135 (2021).
-
Riou, C. et al. Sci. Transl. Med. 14, eabj6824 (2022).
-
Meng, B. et al. Cell Rep. 35, 109292 (2021).
-
Liu, Y. et al. Nature 602, 294–299 (2022).
-
Carreño, J. M. et al. EBioMed. 73, 103626 (2021).
-
West, A. P. et al. Nat. Commun. 12, 4886 (2021).
-
McCarthy, K. R. et al. Science 371, 1139–1142 (2021).
-
Haslwanter, D. et al. mBio 12, 02473–21 (2021).
-
Wibmer, C. K. et al. Nat. Med. 27, 622–625 (2021).
-
Cao, Y. et al. Nature 602, 657–663 (2022).
-
Starr, T.N. et al. Preprint at BioRxiv https://doi.org/10.1101/2022.02.24.481899 (2022).
-
Iketani, S. et al. Nature 604, 553–556 (2022).
-
Chen, R. E. et al. Nat. Med. 27, 717–726 (2021).
-
Wang, Z. et al. Nature 592, 616–622 (2021).
-
Starr, T. N. et al. Science 371, 850–854 (2021).
-
Weisblum, Y. et al. eLife 9, e61312 (2020).
-
Garrett Rappazzo, C. et al. Science 371, 823–829 (2021).
-
Deng, X. et al. Cell 184, 3426–3437 (2021).
-
Harvey, W. T. et al. Nat. Rev. Microbiol. 19, 409–424 (2021).
-
Motozono, C. et al. Cell Host Microbe 29, 1124–1136 (2021).
-
Wang, M. et al. Emerg. Microbes Infect. 11, 18–29 (2022).
-
Tada, T. et al. Front. Immunol. 13, 797589 (2022).
-
Starr, T. N. Cell 182, 1295–1310.e20 (2020).
-
Liu, Z. et al. Cell Host Microbe 29, 477–488 (2021).
-
Greaney, A. J. et al. Cell Host Microbe 29, 463–476.e6 (2021).
-
Andreano, E. et al. Proc. Natl Acad. Sci. USA 118, e2103154118 (2021).
-
Greaney, A. J. et al. Nat. Commun. 12, 4196 (2021).
-
Li, Q. et al. Cell 184, 2362–2371.e9 (2021).
-
Collier, D. A. et al. Nature 593, 136–141 (2021).
-
Volz, E. et al. Cell 184, 64–75.e11 (2021).
-
Hou, Y. J. et al. Science 370, 1464–1468 (2020).
-
Escalera, A. et al. Cell Host Microbe 30, 373–387.e7 (2022).
-
Baum, A. et al. Science 369, 1014–1018 (2020).
-
Lubinski, B. et al. iScience 25, 103589 (2022).
-
Liu, Y. et al. Cell Rep. 39, 110829 (2022).
-
Saito, A. et al. Nature 602, 300–306 (2022).
Acknowledgements
We acknowledge scientists, public health professionals and Ministries of Health across the world for early generation and sharing of data on SARS-CoV-2 variants.
Author information
Authors and Affiliations
Contributions
L.S., M.P.G.K. and A.A. drafted the initial manuscript. A.v.G, B.B.O.M., L.J.A.-R., X.A., E.A. B.N.A, H.A.C., A.B., W.S.B., J.N.B.,L.C., M. Chand, M. Chen, A.C., T.d.O., C.D., J.D., P.E., I.E.M., A.F., S.G., E.G., R.G., B.L. Haagmans, B.L. Herring, S.S.I., Z.K., M.K., R.J.K., J.A.L., Y.-S.L., G.M.L., M.M, S.M., J.M.-R., N.M.M., V.M., K.N., D.-Y.O., B.I.P., T.P.P., M.P., Z.P., L.L.M.P., A.R., J.S., Y.S., M.M.S., S.K.T., E.M.V., V.T., S.v.d.W., S.B., M.D.P. and M.D.V.K. critically read and contributed to the manuscript. N.W., L.T. and S.R. produced the figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Subissi, L., von Gottberg, A., Thukral, L. et al. An early warning system for emerging SARS-CoV-2 variants. Nat Med (2022). https://ift.tt/kod3QvG
-
Published:
-
DOI: https://ift.tt/kod3QvG
Health - Latest - Google News
May 30, 2022 at 04:50PM
https://ift.tt/UpFNcqy
An early warning system for emerging SARS-CoV-2 variants - Nature.com
Health - Latest - Google News
https://ift.tt/Bf2j3DR
Bagikan Berita Ini














0 Response to "An early warning system for emerging SARS-CoV-2 variants - Nature.com"
Post a Comment