A recent study conducted by a team of scientists in the Netherlands and the USA has revealed that while severely affected hospitalized coronavirus disease 2019 (COVID-19) patients and vaccinated individuals are capable of neutralizing the B.1.1.7, B.1.351, and P.1 variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a significant proportion of non-hospitalized patients with less severe COVID-19 remain susceptible to these viral variants.
A preprint version of the study is available on the medRxiv* server, while the article undergoes peer review.
Background
With 171.3 million confirmed COVID-19 cases and 3.5 million deaths, the COVID-19 pandemic has become the largest pandemic in modern history. The frequent emergence of novel SARS-CoV-2 variants is continuously putting the global population under serious threat. Because of significantly increased infectivity and immune evasion ability, the World Health Organization (WHO) has designated some viral variants as the Variants of Concern (VOCs).
Among recognized VOCs, B.1.1.7, B.1.351, and P.1 have been identified first in the UK, South Africa, and Brazil, respectively. Soon after their emergence, these variants have started spreading globally, and cases with these variants have been detected in 132, 82, and 52 countries, respectively.
The N501Y mutation in the spike receptor-binding domain (RBD) is the common feature in all three VOCs. This mutation is known to increase the RBD affinity for human angiotensin-converting enzyme 2 (ACE2), explaining the increased infectivity of these VOCs. In addition, the E484K mutation found in B.1.351 and P.1 variants is known to facilitate viral escape from antibody-mediated neutralization.
In the current study, the scientists have compared the immune escape abilities of B.1.1.7, B.1.351, and P.1 variants. Specifically, they have investigated whether these VOCs can escape neutralization by monoclonal therapeutic antibodies and polyclonal antibodies derived from COVID-19 patients and vaccinated individuals.
Study design
For antibody testing, serum samples were obtained from 69 COVID-19 patients 4 – 6 weeks after the symptom onset. An additional set of serum samples was collected from 50 vaccinated healthcare workers 4 weeks after the 2nd vaccine dose. The participants received the mRNA-based COVID-19 vaccine developed by Pfizer/BioNTech.
The spike proteins of wild-type SARS-CoV-2 and three VOCs were generated and used to measure the binding antibody responses of collected serum samples using multiplex protein microarray.
To determine the neutralizing ability of sera, lentiviral-based pseudoviruses of the wild-type virus and three VOCs were utilized.
Important observations
The study findings revealed that compared to the wild type SARS-CoV-2, the spike binding ability of convalescent sera reduced by 2.4-fold, 3-fold, and 4-fold for the B.1.1.7, B.1.351, and P.1 variants, respectively. Moreover, a highly significant difference in spike-binding antibody levels was observed between severely affected hospitalized COVID-19 patients and mildly affected non-hospitalized COVID-19 patients. A comparable level of binding antibody was observed in vaccinated individuals and hospitalized patients, which was 4 – 11 times higher than non-hospitalized patients. All these observations were consistent for all tested VOCs.
Regarding virus neutralization, about 96% of convalescent sera and 100% of vaccinated sera exhibited complete neutralizing ability against the wild-type virus. However, all tested sera showed a significant reduction in neutralization potency against the VOCs. Specifically, non-hospitalized patients, hospitalized patients, and vaccinated individuals showed 4-fold, 7-fold, and a 5-fold reduction in neutralizing titers against the B.1.351 variant, respectively.
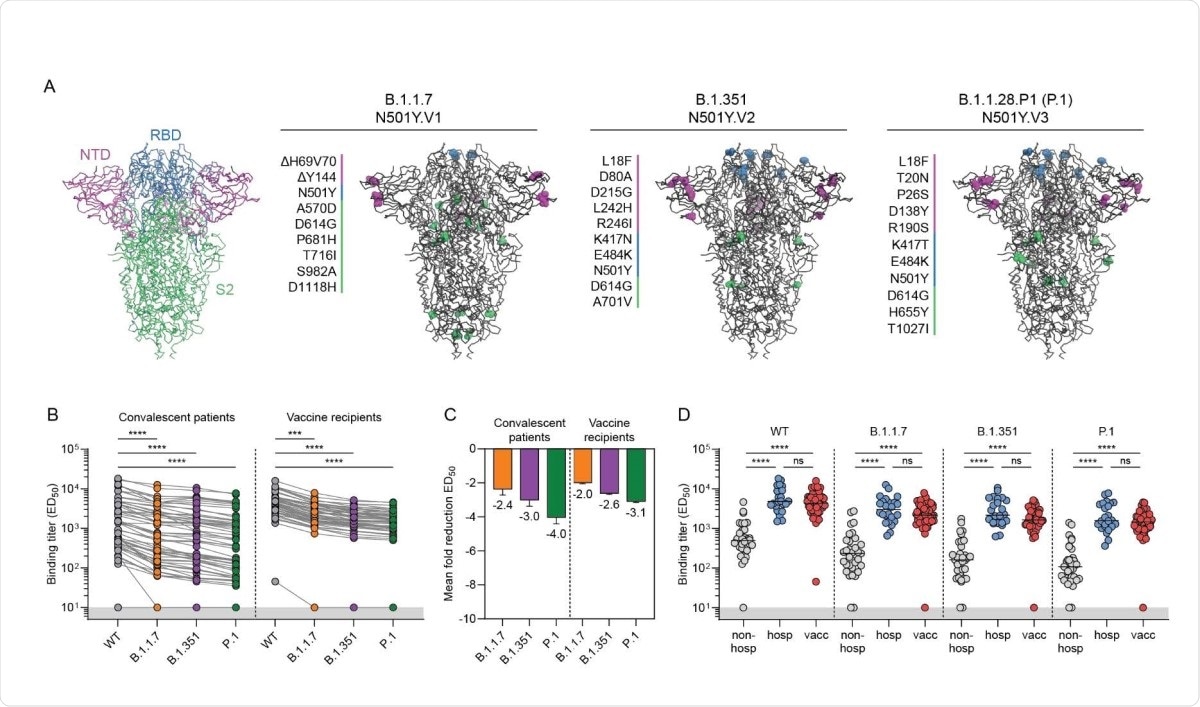
While 100% of non-hospitalized patients showed neutralizing titers against the wild-type virus and B.1.1.7 variant, about 39% and 34% of them failed to neutralize the B.1.351 and P.1 variants, respectively. In contrast, all hospitalized patients and vaccinated individuals retained at least some neutralization ability against the VOCs. Overall, these observations indicate that a high neutralizing titer against the wild-type virus is predictive for cross-neutralization of VOCs.
Regarding monoclonal antibodies, preclinically tested anti-RBD and anti-NTD (N-terminal domain) antibodies showed significantly reduced binding against the B.1.351 and P.1 variants compared to that against the wild-type virus. However, some SARS-CoV cross-reactive anti-RBD monoclonal antibodies retained similar binding kinetics to all tested VOCs. The mutational analysis of VOCs revealed that the E484K mutation, together with other mutations such as K417N and K417T, had the highest impact on the binding of monoclonal anti-RBD antibodies.
Regarding virus neutralization, only 5 out of 11 tested anti-RBD monoclonal antibodies retained neutralizing ability against the B.1.351 and P.1 variants. However, both anti-NTD antibodies failed to neutralize the B.1.351 variant. Importantly, SARS-CoV cross-reactive monoclonal antibody successfully retained its ability to neutralize all tested VOCs. This indicates that this antibody targets a highly conserved epitope that is not impacted by the mutations found in these VOCs.
Study significance
The study findings reveal that despite having a low binding and neutralizing titers, hospitalized patients with severe COVID-19 and vaccinated individuals are capable of neutralizing the B.1.1.7, B.1.351, and P.1 variants of SARS-CoV-2. However, mildly affected non-hospitalized COVID-19 patients fail to induce sufficient neutralizing titers against these VOCs. Among tested VOCs, B.1.351 variant exhibits the highest resistance to antibody-mediated neutralization.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Health - Latest - Google News
June 03, 2021 at 02:55AM
https://ift.tt/3fIK1V6
Novel SARS-CoV-2 variants partially resistant to infection- and vaccine-induced immunity, study finds - News-Medical.Net
Health - Latest - Google News
https://ift.tt/2zrj9Ud
Bagikan Berita Ini














0 Response to "Novel SARS-CoV-2 variants partially resistant to infection- and vaccine-induced immunity, study finds - News-Medical.Net"
Post a Comment